X = dat[which(dat[,1] == 4), -1]
dim(X)[1] 652 256In the next series of tasks, we implement image reconstruction with HWD dataset.
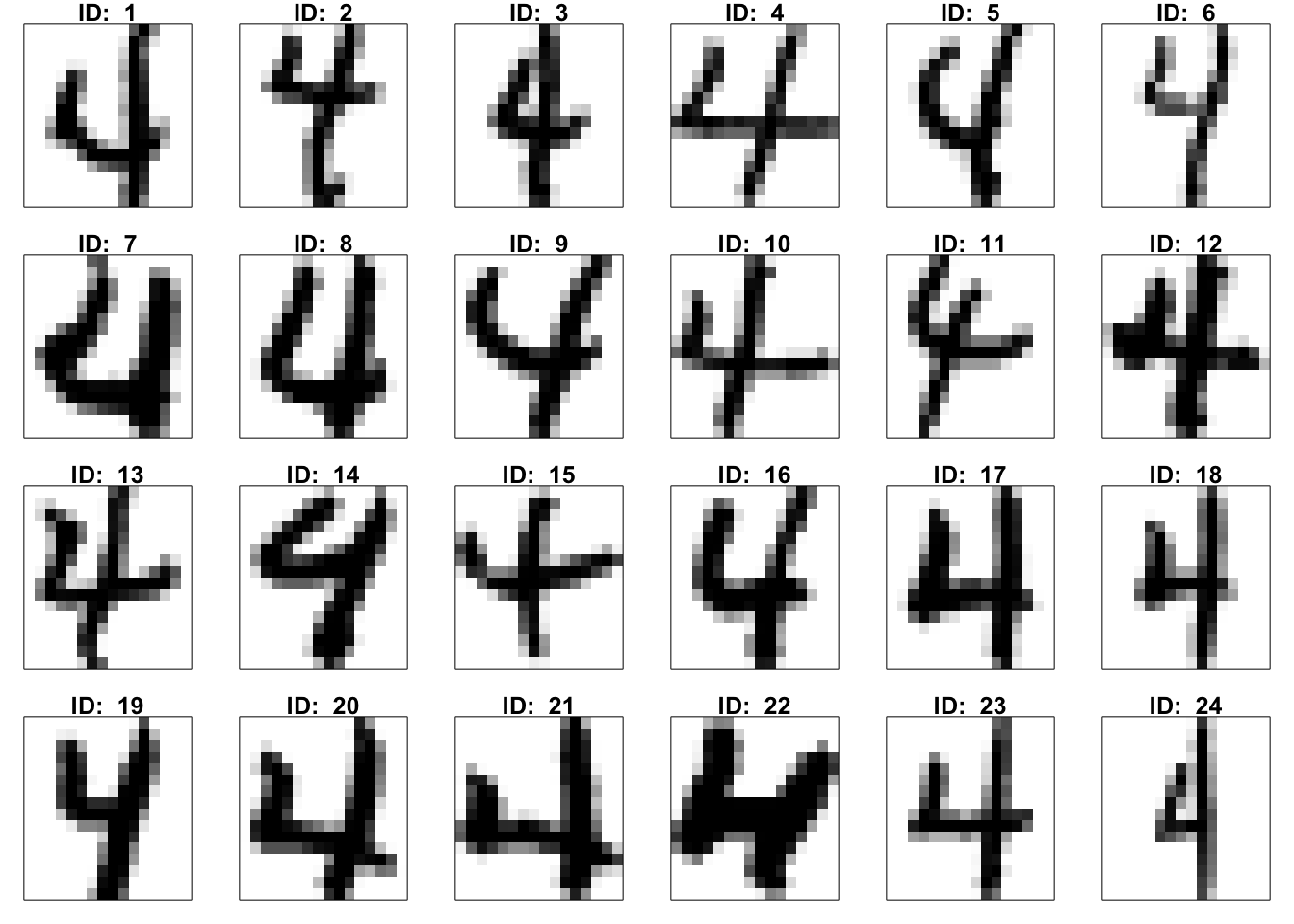
Task 1.1: Choose your favorite number. Use the function ‘image’ to display the first 24 cases. Below is an example of the usage method for the function image(). In the following R chunk, the first two lines are about color settings. You can also try your favorite color palette. In the third line, X represents all images of your favorite number, which refers to the part of the Dat matrix in R space excluding the first column. The last line generates the plot of the 8th image in X.
colors = c('white','black')
cus_col <- colorRampPalette(colors=colors)
temp = matrix(X[8,256:1],16,16,byrow=T)[,16:1]
image(t(temp), col=cus_col(256), frame = T, axes = F)Choose all the images of my favorite number:
Visualize the first 24 images in the dataset by using function image()
colors = c('white','black')
cus_col <- colorRampPalette(colors=colors)
par(mfrow = c(4,6), mar = rep(1,4))
for(i in 1:24){
temp= matrix(X[i,256:1],16,16,byrow=T)[,16:1]
image(t(temp), col=cus_col(256), frame = T, axes = F)
title(paste("ID: ", i))
}
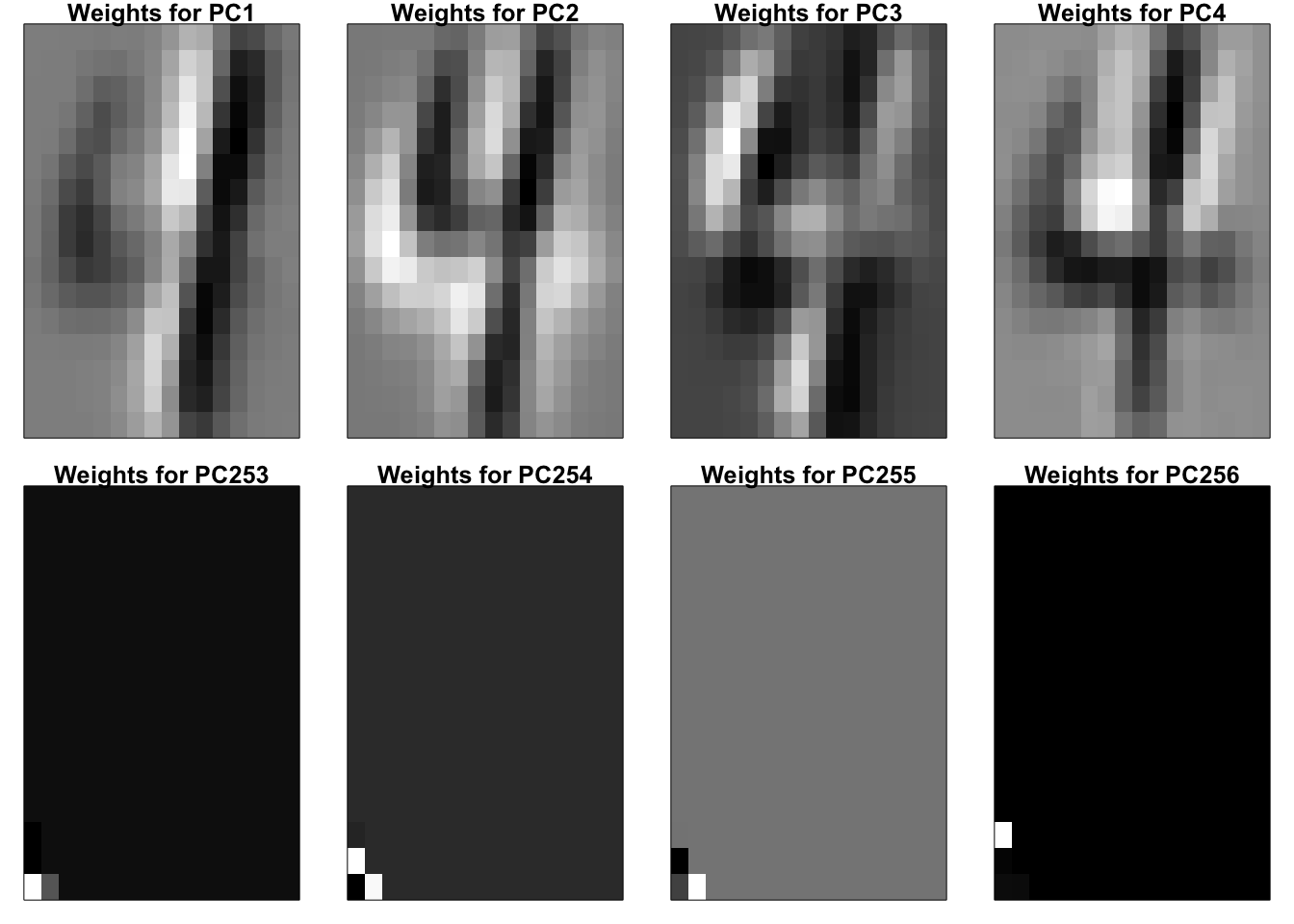
Task 1.2: Do PCA on your data set. Display the first 4 and the last 4 principal vectors (eigenvectors) as \(16 \times 16\) images.
To do PCA, we first calculate the covariance matrix of X, then do eigenvalue decomposition on the contrivance matrix.
All the PC weights are saved in W; each column is a set of weights for calculating the corresponding new variable, PC. Next, we display the first 4 and the last 4 columns of W as images.
par(mfrow = c(2,4), mar = rep(1,4))
for(i in c(1:4, 253:256)){
temp = matrix(W[256:1,i],16,16,byrow=T)[,16:1]
image(t(temp), col=cus_col(256), frame = T, axes = F)
title(paste0("Weights for PC", i))
}
Task 1.3: Calculate all the PCs for all the images of your favorite number.
If you are familiar with matrix calculations, then you can get the new data matrix directly
Otherwise, you can calculate all the PCs for all the image by a double for lop.
Task 1.4: Image reconstruction. Reconstruct an image in your data set by its first 30, 60, 100, 150, and 200 principal components separately. Put the original image and five approximated images in one plot. For each approximated image, calculate and report the mean square errors.
This also can be done by matrix calculation. However, we will do it by for loop first since it is more understandable. Next, I will reconstruct the 8th image of number 4.
k = 150 # we use k PCs to reconstruct the image
x_hat = Z[8,1]*W[,1]
for(i in 2:k){
x_hat = x_hat + Z[8,i]*W[,i]
}
# vis
par(pin = c(4, 4))
temp = matrix(x_hat[256:1],16,16,byrow=T)[,16:1]
image(t(temp), col=cus_col(256), frame = T, axes = F)
The approximation error is
You can get the same results by matrix multiplication.
Build a classifier based on the first two principal components (PCs) to classify the images of digits 5 and 6. The following procedure is suggested to solve this task.
Step 1:
Step 2:
Step 3:
S = cov(X_tr)
W = eigen(S)$vectors
Z_tr = X_tr%*%W[,1:2]
Z_te = X_te%*%W[,1:2]
par(mfrow = c(1,2))
plot(Z_tr, col = y_tr)
plot(Z_te, col = y_te)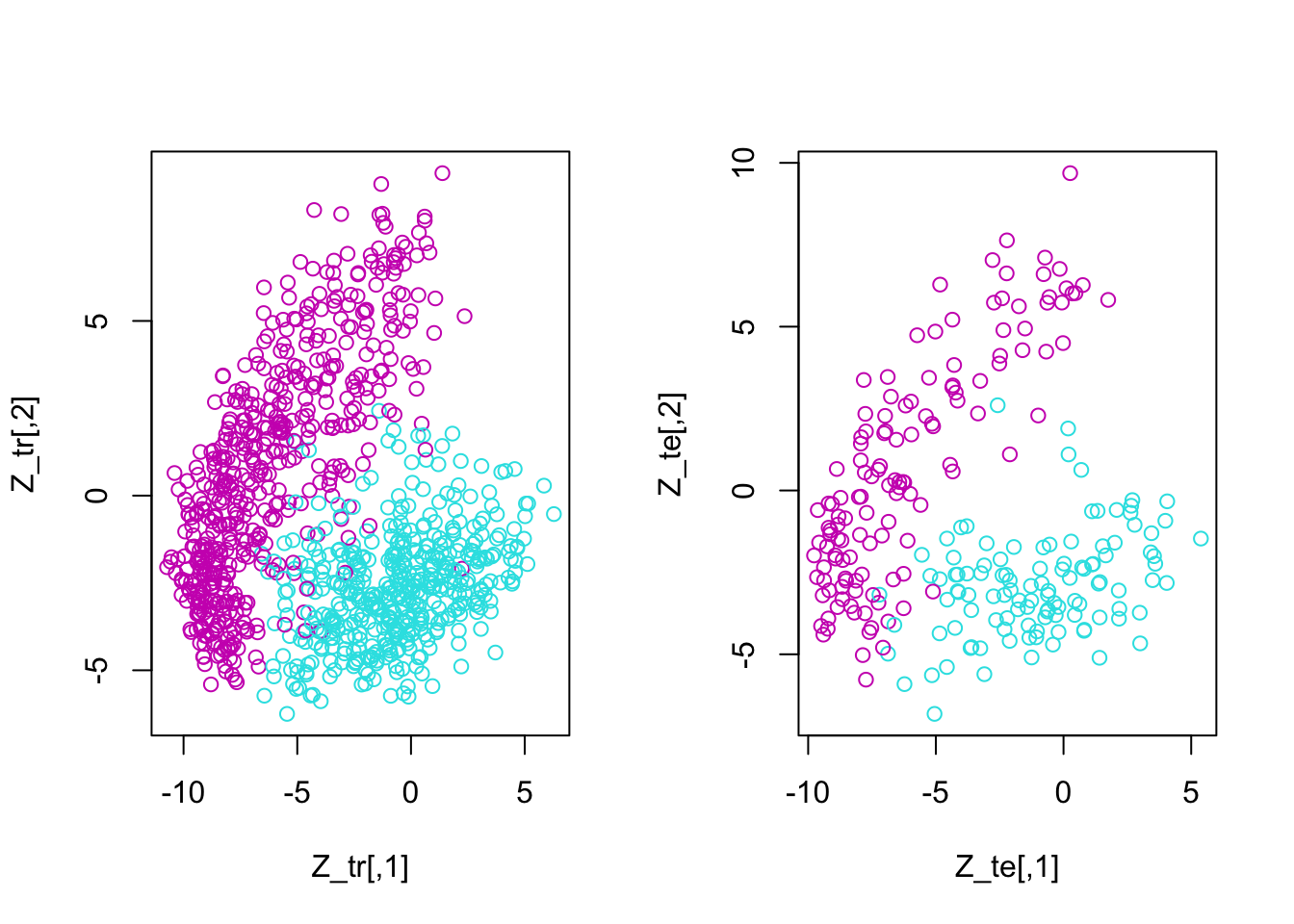
Step 4:
y z1 z2
1 1 -5.2767511 -4.9867831
2 0 -8.3540556 -4.3457323
3 0 -2.1933222 3.4651025
4 0 -2.7204058 -0.3158707
5 1 -0.5886785 -3.9040087
6 0 -7.7362370 -0.4245627m = glm(y~z1+z2, family = binomial(), data = d_tr)
w = m$coefficients
par(mfrow = c(1,2))
plot(Z_tr, col = y_tr)
abline(-w[1]/w[3], -w[2]/w[3])
plot(Z_te, col = y_te)
abline(-w[1]/w[3], -w[2]/w[3])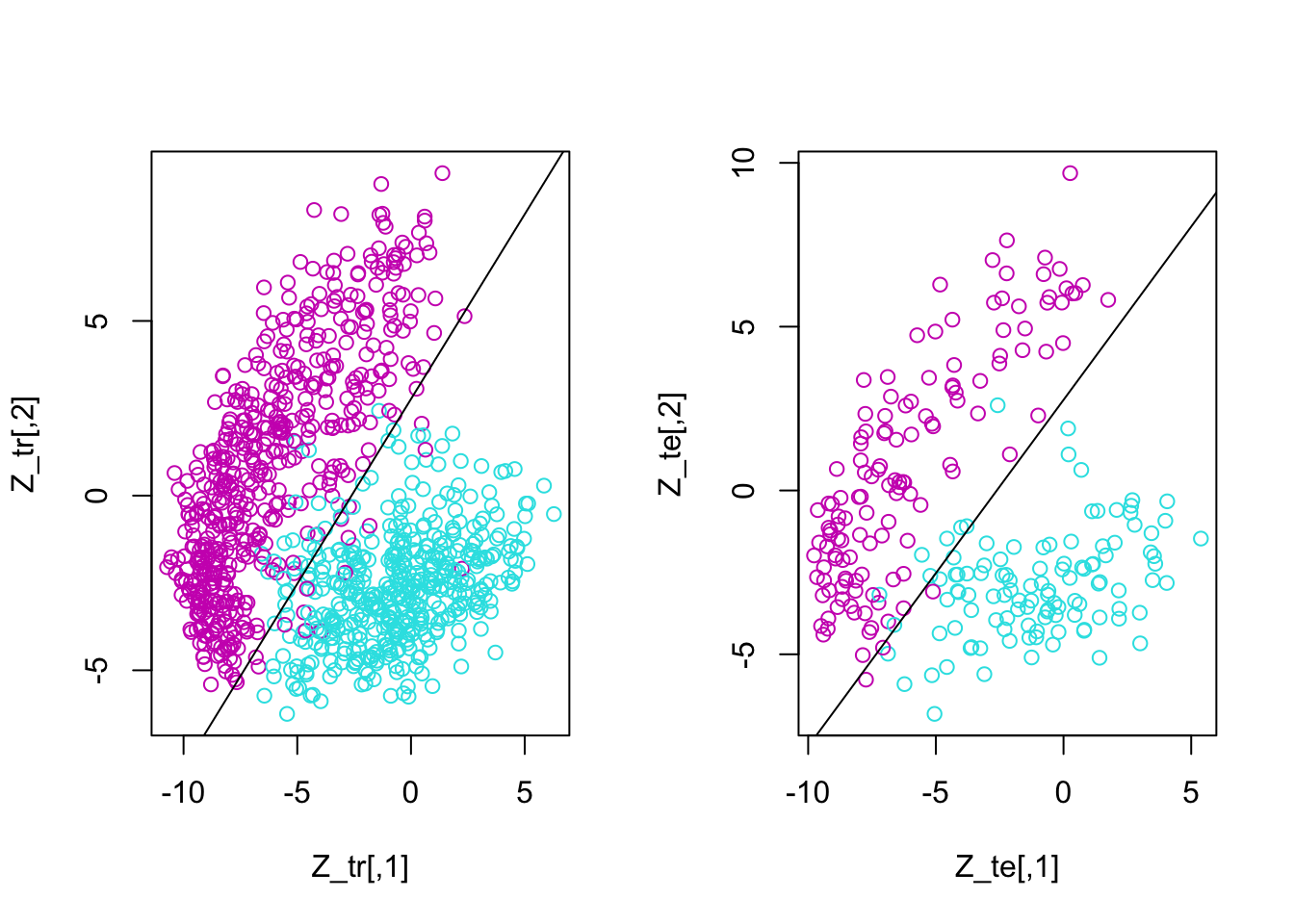
Step 5:
d_te = data.frame(y = ifelse(y_te == 5, 1, 0), z1 = Z_te[,1], z2 = Z_te[,2])
res = predict(m, d_te, type = "response")
y_pre = ifelse(res > 0.5, 1, 0)
mean(y_pre == d_te$y)[1] 0.954918
y_pre 0 1
0 129 8
1 3 104Can you achieve this level of accuracy using only two variables from the original dataset to build the classifier? Think about it.
